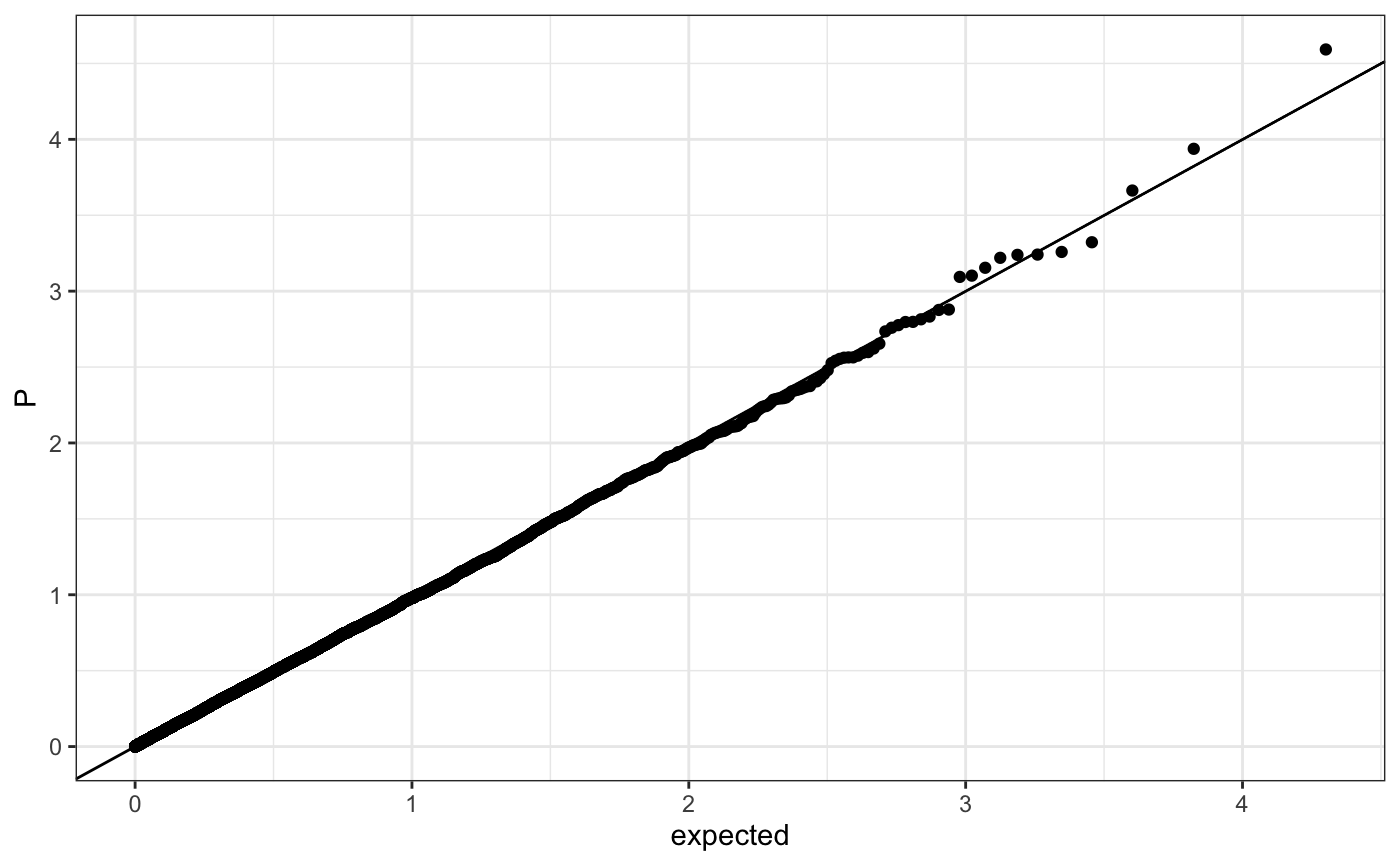
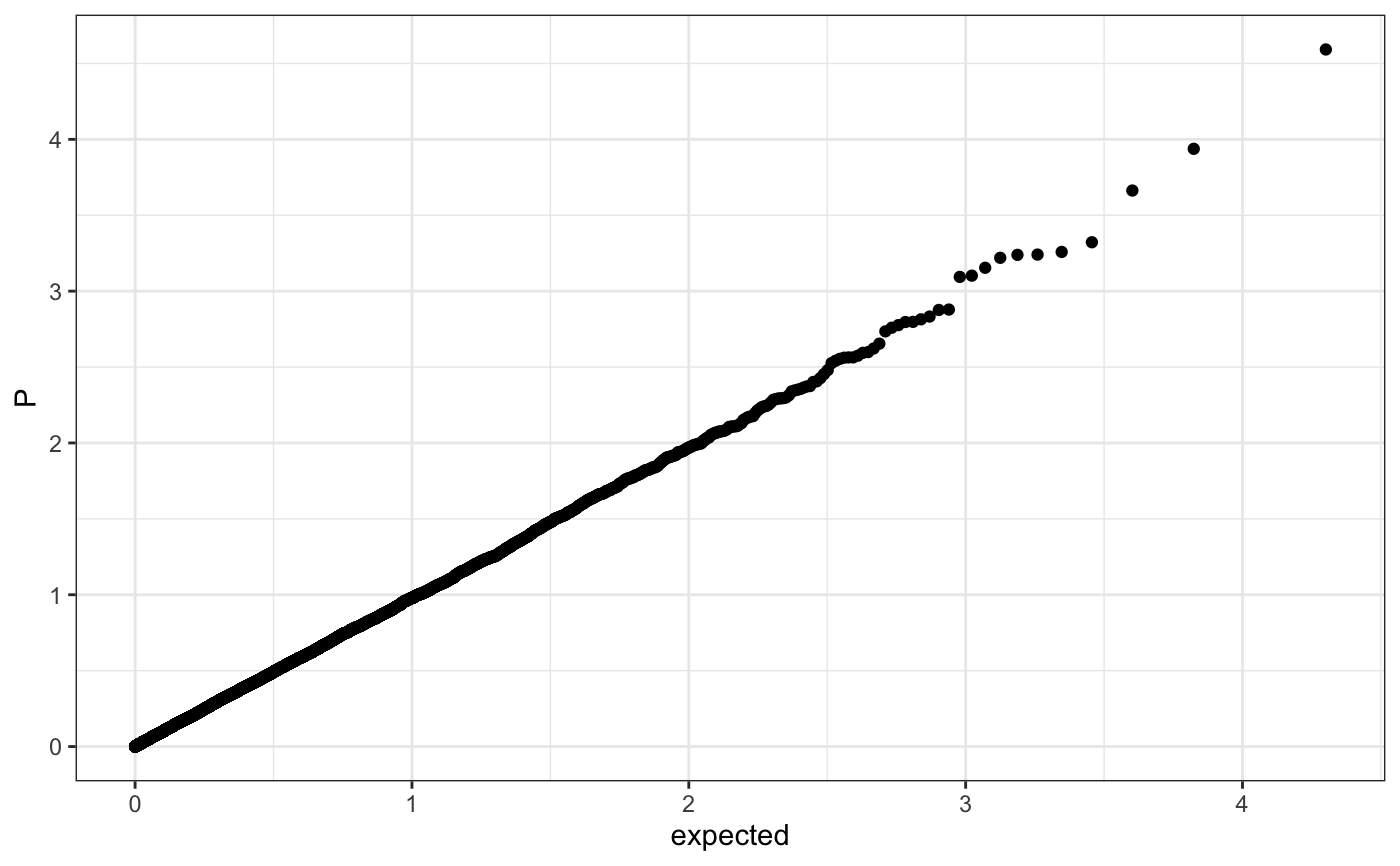
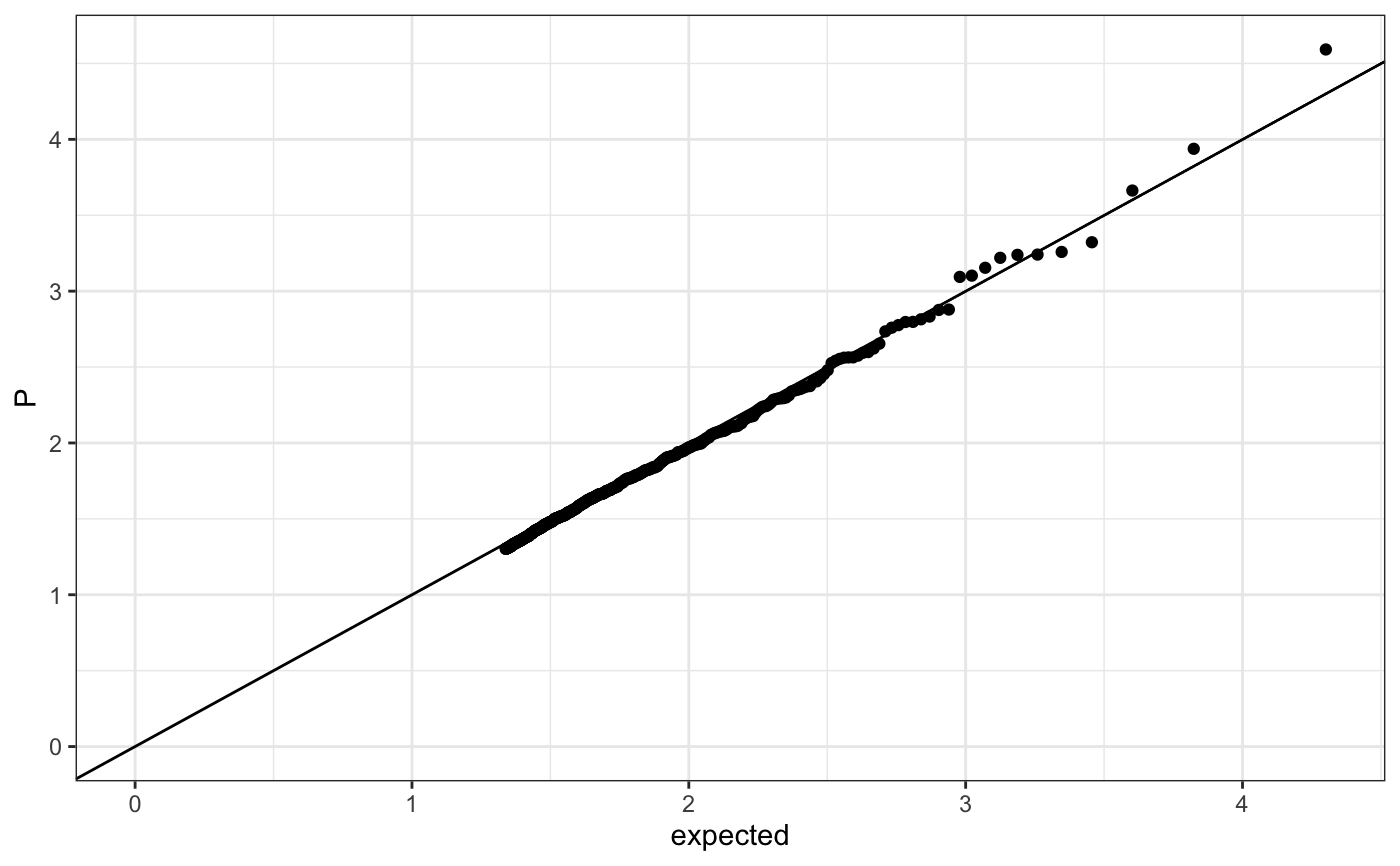
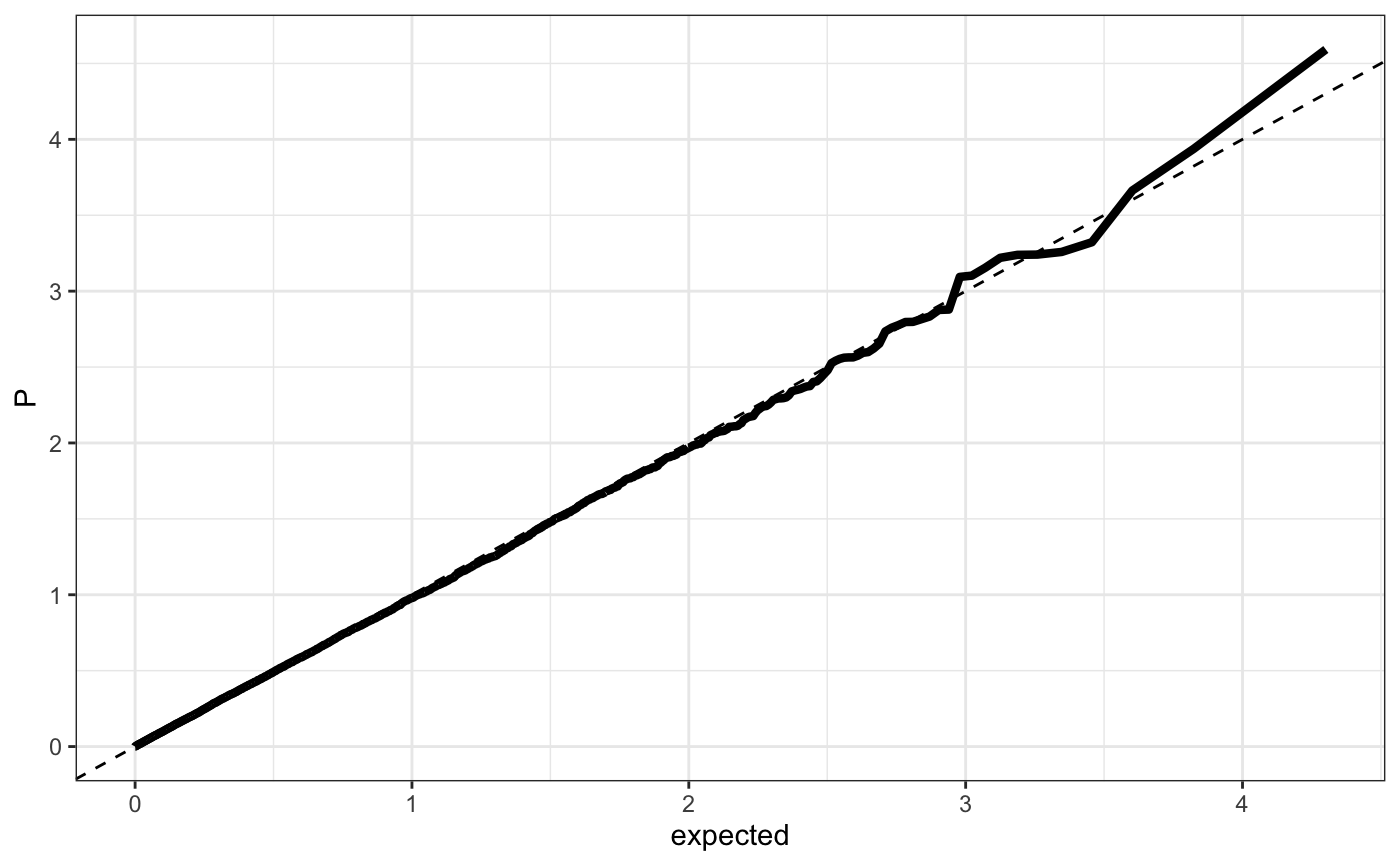
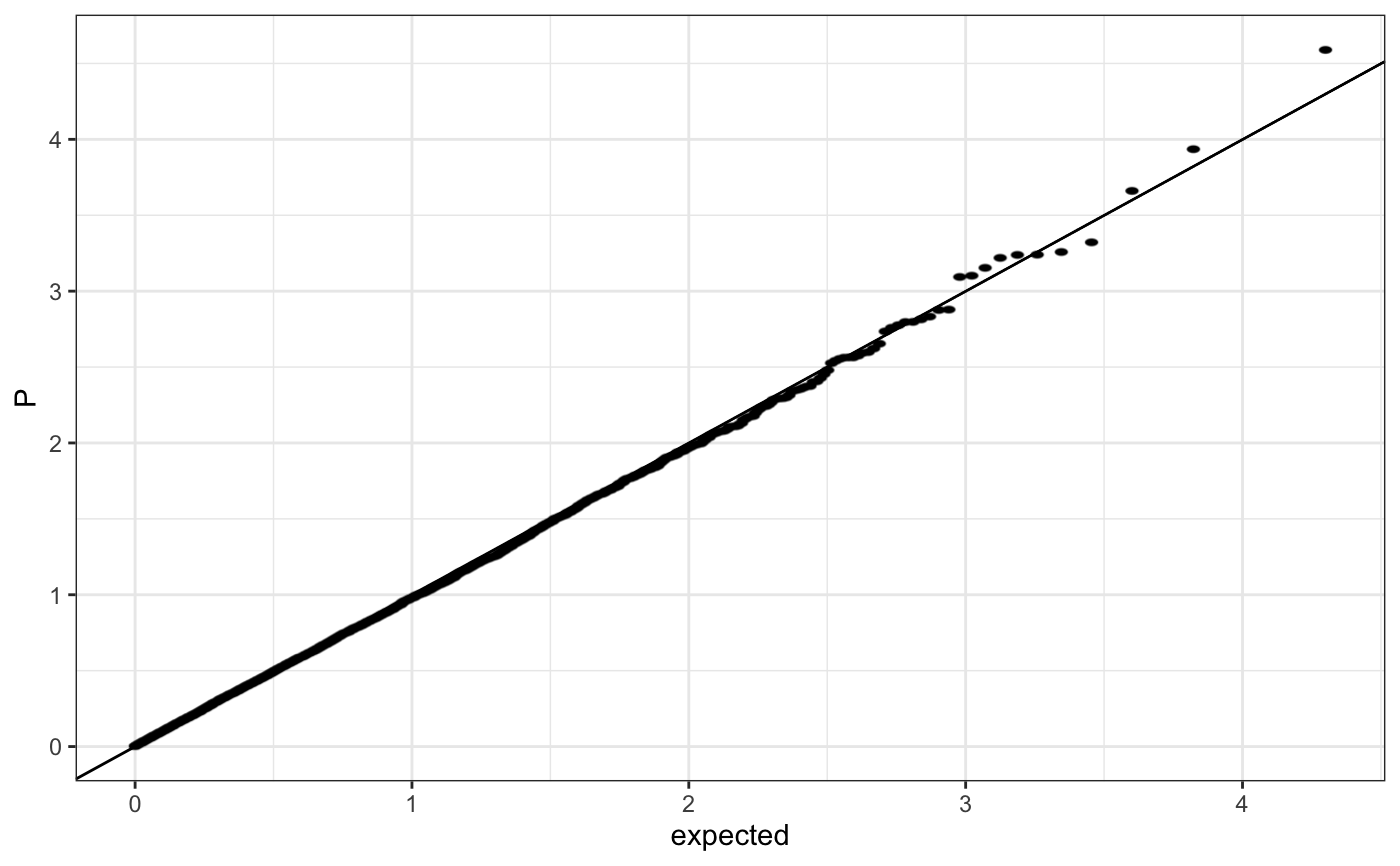
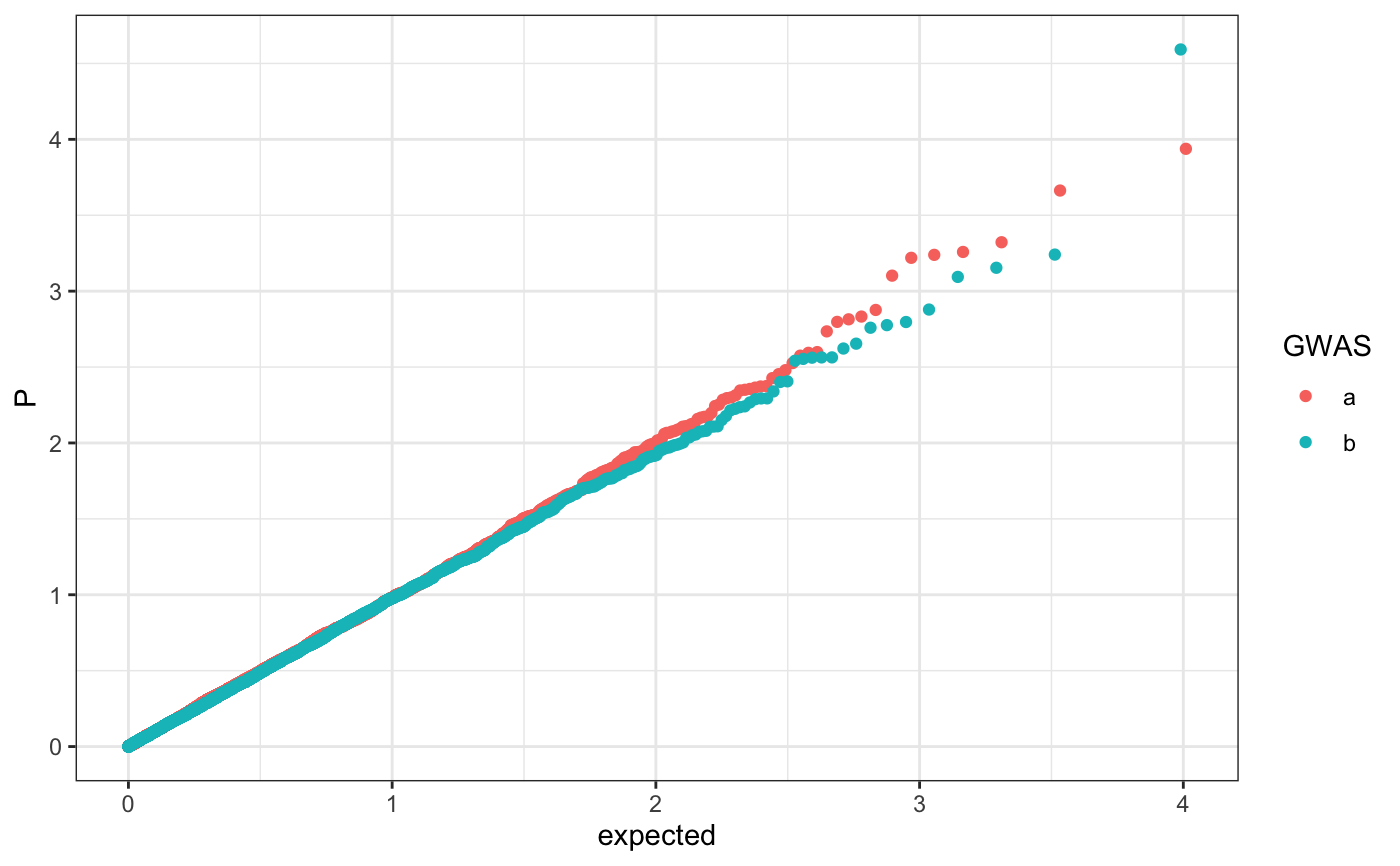
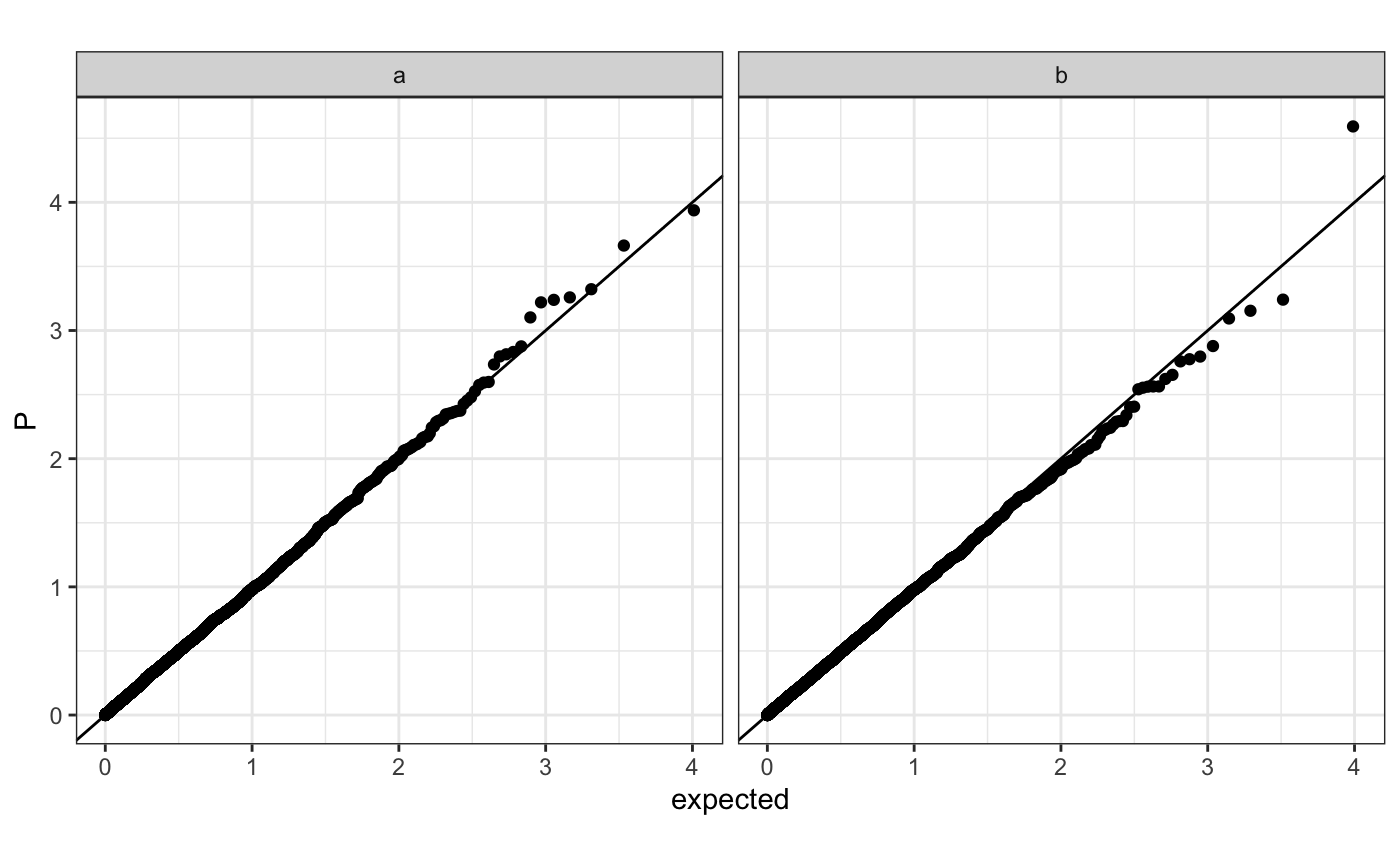
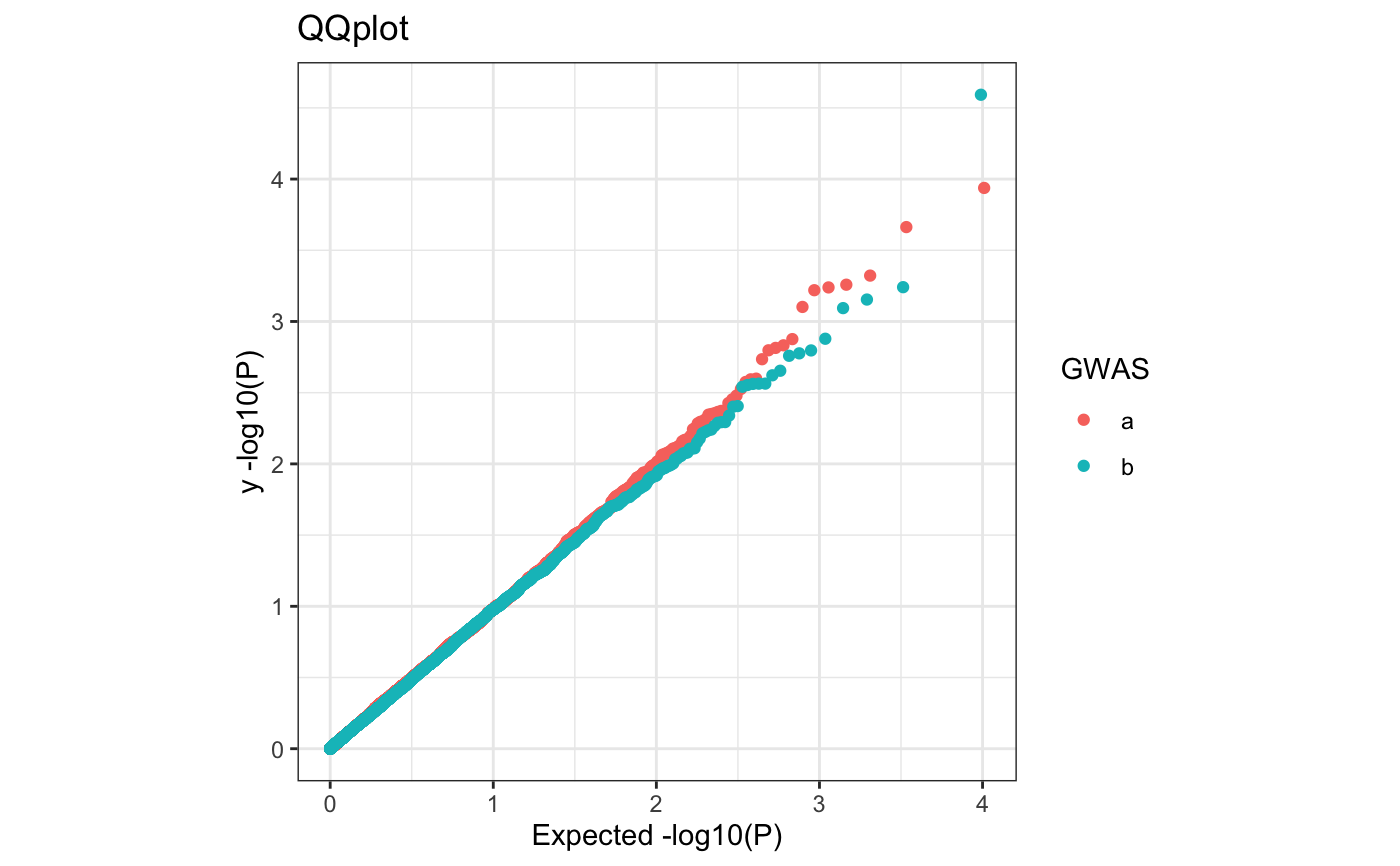
Q-Q plot
stat_gwas_qq.RdQuantile-quantile plot to compare the p-values of a GWAS to a uniform distribution.
stat_gwas_qq(mapping = NULL, data = NULL, geom = "point", position = "identity", na.rm = FALSE, show.legend = NA, inherit.aes = TRUE, y.thresh = NULL, ...) geom_gwas_qq(mapping = NULL, data = NULL, geom = "point", position = "identity", na.rm = FALSE, show.legend = NA, inherit.aes = TRUE, y.thresh = NULL, ...)
Arguments
| mapping | Set of aesthetic mappings created by |
|---|---|
| data | The data to be displayed in this layer. There are three options: If A A |
| geom |
|
| position | Position adjustment, either as a string, or the result of a call to a position adjustment function. |
| na.rm | If |
| show.legend | logical. Should this layer be included in the legends?
|
| inherit.aes | If |
| y.thresh | same scale as y (e.g. 0.05), y <= y.thresh AFTER computing expected |
| ... | Other arguments passed on to |
Details
Alternatively, use stat_qq, that works for
all kinds of distributions, together with mlog_trans.
Note
Plotting several thousand points might take time. If you want to speed
things up use stat_gwas_qq_hex.
See also
Examples
require(ggplot2) n.sample <- 10000 df <- data.frame(P = runif(n.sample), GWAS = sample(c("a", "b"), n.sample, replace = TRUE )) theme_set(theme_bw()) ## default (qp <- ggplot(df, aes(y = P)) + stat_gwas_qq() + geom_abline(intercept = 0, slope = 1))## show only p-values above a cerain threshold ggplot(df, aes(y = P)) + stat_gwas_qq(y.thresh = 0.05) + geom_abline(intercept = 0, slope = 1) + xlim(0, NA) + ylim(0, NA)## plot a line instead ggplot(df, aes(y = P)) + stat_gwas_qq(geom = "line", size = 1.5) + geom_abline(intercept = 0, slope = 1, linetype = 2)## plot efficiently ggplot(df, aes(y = P)) + stat_gwas_qq(geom = ggrastr:::GeomPointRast) + geom_abline(intercept = 0, slope = 1)## Group and color points according to GWAS (qp <- ggplot(df, aes(y = P)) + stat_gwas_qq(aes( group = GWAS, color = GWAS )))## facet ggplot(df, aes(y = P)) + facet_wrap(~GWAS) + stat_gwas_qq() + geom_abline(intercept = 0, slope = 1) + theme(aspect.ratio = 1)## adding nice labels, square shape ## identical limits (meaning truely square) qp + theme(aspect.ratio = 1) + ## square shaped expand_limits(x = -log10(max(df$P)), y = -log10(max(df$P))) + ggtitle("QQplot") + xlab("Expected -log10(P)") + ylab("y -log10(P)")## group library(GWAS.utils) ## devtools::install_github("sinarueeger/GWAS.utils") data("giant") ?giant ## generate two groups giant <- giant %>% dplyr::mutate(gr = dplyr::case_when( BETA <= 0 ~ "Neg effect size", BETA > 0 ~ "Pos effect size" )) ggplot(data = giant, aes(y = P, group = gr, color = gr)) + stat_gwas_qq() + geom_abline(intercept = 0, slope = 1)